human_annotations_json = '/content/drive/MyDrive/2024-01-04-Text-Annotation-Sample.json' # @param {type:"string"}
import json
with open(human_annotations_json, 'r') as f:
j = f.read()
exported_annotations = json.loads(j)
import pandas as pd
from tqdm.notebook import tqdm
def process_result(result, coder, md):
value_type = result.get('type', "")
metadata = {
**md,
"coder": coder,
"from_name": result.get('from_name', "")
}
annotations = []
if value_type == "choices":
choices = result['value'].get('choices', [])
for choice in choices:
r = {**metadata}
r['value'] = choice
annotations.append(r)
elif value_type == "taxonomy":
taxonomies = result['value'].get('taxonomy', [])
for taxonomy in taxonomies:
if len(taxonomy) > 1:
taxonomy = " > ".join(taxonomy)
elif len(taxonomy) == 1:
taxonomy = taxonomy[0]
r = {**metadata}
r['value'] = taxonomy
annotations.append(r)
return annotations
all_annotations = []
for data in tqdm(exported_annotations):
annotations = data.get("annotations")
metadata = {
**data.get("data")
}
for annotation in annotations:
coder = annotation['completed_by']['id']
results = annotation.get("result")
if results:
for result in results:
all_annotations.extend(process_result(result, coder, metadata))
else:
print("Skipped Missing Result")
all_annotations_df = pd.DataFrame(all_annotations)Read the LabelStudio Annotations from file
Enter the location (e.g. on Google Drive) of your exported json file and run this cell. It loads the annotations in a long format into the variable all_annotations_df.
This version does not handle reviews!
This version has been tested with binary classifications. Other types of nominal data should work out of the box for interrater agreement, model evaluation needs some refactoring!
# Check the dataframe
all_annotations_df.head()| Text | uuid | Filename | Identifier | coder | from_name | value | |
|---|---|---|---|---|---|---|---|
| 0 | CDU #SOFORTPROGRAMM MITTELSTANDSPAKET WIR MACH... | 3733 | cdu/2021-09-13_15-01-28_UTC.jpg | CTxB6cMKe2U | 13724 | Positioning | True |
| 1 | CDU #SOFORTPROGRAMM MITTELSTANDSPAKET WIR MACH... | 3733 | cdu/2021-09-13_15-01-28_UTC.jpg | CTxB6cMKe2U | 13724 | Call to Action | False |
| 2 | CDU #SOFORTPROGRAMM MITTELSTANDSPAKET WIR MACH... | 3733 | cdu/2021-09-13_15-01-28_UTC.jpg | CTxB6cMKe2U | 13724 | Documentation | False |
| 3 | CDU #SOFORTPROGRAMM MITTELSTANDSPAKET WIR MACH... | 3733 | cdu/2021-09-13_15-01-28_UTC.jpg | CTxB6cMKe2U | 13724 | OCR | False |
| 4 | CDU #SOFORTPROGRAMM MITTELSTANDSPAKET WIR MACH... | 3733 | cdu/2021-09-13_15-01-28_UTC.jpg | CTxB6cMKe2U | 7379 | Positioning | True |
Contingency Table
We select one coding in this step (e.g. Positioning) and create a contingency table where each annotated item (text, image) occupies one row and each coder one column. The identifier should be set to a unique column, like uuid or Filename.
Enter the from_name for the variable you’re interested in at the moment. (Refer to your LabelStudio Interface for the right from_name).
import pandas as pd
import numpy as np
from_name = 'Positioning'
identifier = 'uuid'
filtered_df = all_annotations_df[all_annotations_df['from_name'].str.contains(from_name, case=False)]
def to_bool(val):
if isinstance(val, bool):
return val
if isinstance(val, str):
return val.lower() == "true"
return bool(val)
values = filtered_df['value'].unique()
identifier_values = filtered_df[identifier].unique()
contingency_matrix = pd.crosstab(filtered_df[identifier], filtered_df['coder'], values=filtered_df['value'], aggfunc='first')
contingency_matrix = contingency_matrix.reindex(identifier_values)# Let's take a look at the contigency table. We refer to each coder using a pseudonymous number.
contingency_matrix.head()| coder | 7107 | 7379 | 10506 | 13724 |
|---|---|---|---|---|
| uuid | ||||
| 3733 | NaN | True | True | True |
| 4185 | False | True | True | NaN |
| 530 | NaN | True | False | True |
| 2721 | NaN | False | False | False |
| 3384 | NaN | False | False | False |
Calculate Pairwise \(\kappa\)
Let’s calculate Cohen’s Kappa for each coder pair.
from sklearn.metrics import cohen_kappa_score
# Function to calculate Cohen's Kappa for each pair of raters
def calculate_kappa(matrix):
raters = matrix.columns
kappa_scores = []
for i in range(len(raters)):
for j in range(i+1, len(raters)):
rater1, rater2 = raters[i], raters[j]
# Drop NA values for the pair of raters
pair_matrix = matrix[[rater1, rater2]].dropna()
# Skip pairs without overlaps
if len(pair_matrix) > 0:
kappa = cohen_kappa_score(pair_matrix[rater1], pair_matrix[rater2])
kappa_scores.append({
"Coder 1": rater1,
"Coder 2": rater2,
"Kappa": kappa,
"Overlap": len(pair_matrix),
"Coding": from_name
})
return kappa_scores
kappa_scores = calculate_kappa(contingency_matrix)
kappa_scores_df = pd.DataFrame(kappa_scores)# Let's display the pairwise Kappa agreements.
kappa_scores_df| Coder 1 | Coder 2 | Kappa | Overlap | Coding | |
|---|---|---|---|---|---|
| 0 | 7107 | 7379 | 0.469027 | 20 | Positioning |
| 1 | 7107 | 10506 | 0.782609 | 20 | Positioning |
| 2 | 7379 | 10506 | 0.669500 | 50 | Positioning |
| 3 | 7379 | 13724 | 0.888889 | 30 | Positioning |
| 4 | 10506 | 13724 | 0.714286 | 30 | Positioning |
Overall Agreement: Krippendorff’s \(\alpha\)
Next, we calculate the overall agreement using Krippendorffs Alpha. First we need to install the package.
!pip install krippendorffCollecting krippendorff
Downloading krippendorff-0.6.1-py3-none-any.whl (18 kB)
Requirement already satisfied: numpy<2.0,>=1.21 in /usr/local/lib/python3.10/dist-packages (from krippendorff) (1.23.5)
Installing collected packages: krippendorff
Successfully installed krippendorff-0.6.1import krippendorff
import pandas as pd
def convert_to_reliability_data(matrix):
transposed_matrix = matrix.T
reliability_data = []
for _, ratings in transposed_matrix.iterrows():
reliability_data.append(ratings.tolist())
return reliability_data
reliability_data = convert_to_reliability_data(contingency_matrix)
# Calculating Krippendorff's Alpha treating "Unsure" as a distinct category
alpha = krippendorff.alpha(reliability_data=reliability_data, level_of_measurement='nominal')
print("Krippendorff's Alpha:", alpha)Krippendorff's Alpha: 0.6977687626774848Majority Decision
One approach to assess the quality of machine labelled data is the comparison between machine-generated labels and human-generated labels, commonly known as “gold standard” labels. This process is called “label agreement” or “inter-rater agreement” and is widely used in various fields, including natural language processing, machine learning, and computational social science.
We are going to use create a majority_decision column using the human annotations: We have chosen an uneven number of annotators in order to find a majority for each label. First, we are going to create a contingency table (or matrix), then we can determine the majority decision.
import numpy as np
import pandas as pd
# Each row represents an item, and each column a decision from a different annotator.
# Step 1: Find the mode (most common decision) for each row
# The mode is used as it represents the majority decision.
# decisions will have the most frequent value in each row, handling ties by keeping all modes.
decisions = contingency_matrix.mode(axis=1)
# Step 2: Extract the primary mode (first column after mode operation)
# This represents the majority decision. If there's a tie, it takes the first one.
majority_decisions = decisions.iloc[:, 0]
##########
## Warning: This part needs some refactoring. Will be updated shortly.
##########
# Step 3: Count the number of non-NaN values (actual decisions) per row, excluding the first column
# Use .iloc[:, 1:] to exclude the first column
# row_non_nan_counts = contingency_matrix.iloc[:, 1:].notnull().sum(axis=1)
# Step 4: Invalidate the majority decision where the number of decisions is insufficient or even
# Majority decisions are only considered valid if there are more than 2 decisions and the number of decisions is odd.
# Define a condition for invalidating rows
# invalid_rows_condition = (row_non_nan_counts < 3) | (row_non_nan_counts % 2 == 0)
# Step 5: Append the majority decision as a new column in the contingency matrix
# This column now represents the aggregated decision from the annotators per item.
contingency_matrix['Majority Decision'] = majority_decisionscontingency_matrix.head()| coder | 7107 | 7379 | 10506 | 13724 | Majority Decision |
|---|---|---|---|---|---|
| uuid | |||||
| 3733 | NaN | True | True | True | True |
| 4185 | False | True | True | NaN | True |
| 530 | NaN | True | False | True | True |
| 2721 | NaN | False | False | False | False |
| 3384 | NaN | False | False | False | False |
Import computational annotations Reading the GPT annotations. Enter the correct file path below. coding_column needs to point to the column with you computational annotations, annotated_identifier to an id / filename that has been passed to LabelStudio project. The annotations are merged with the classifications based on annotated_identifier, in my case uuid.
annotated_file = '/content/drive/MyDrive/2024-01-04-Text-Annotation-Sample.csv'
coding_column = 'Positioning'
annotated_identifier = 'uuid'
annotated_df = pd.read_csv(annotated_file)annotated_df.head()| Unnamed: 0 | Text | Post Type | Positioning | uuid | |
|---|---|---|---|---|---|
| 0 | 54 | Gemeinsam als #eineUnion stehen wir für eine ... | Post | True | 54 |
| 1 | 200 | Wir wünschen allen Schülerinnen und Schüler... | Post | True | 200 |
| 2 | 530 | Fränkisches Essen gibt Kraft: Nach einem Mitt... | Post | False | 530 |
| 3 | 707 | Kleines Zwischenfazit zum #TvTriell. | Post | False | 707 |
| 4 | 899 | In einem paar Tagen sind Wahlen, wichtige Wahl... | Story | False | 899 |
contingency_table = pd.merge(contingency_matrix, annotated_df[['uuid', coding_column]], left_on=identifier, right_on=annotated_identifier, how='left')
contingency_table.rename(columns={coding_column: "Model"}, inplace=True)contingency_table.head()| uuid | 7107 | 7379 | 10506 | 13724 | Majority Decision | Model | |
|---|---|---|---|---|---|---|---|
| 0 | 3733 | NaN | True | True | True | True | True |
| 1 | 4185 | False | True | True | NaN | True | True |
| 2 | 530 | NaN | True | False | True | True | False |
| 3 | 2721 | NaN | False | False | False | False | False |
| 4 | 3384 | NaN | False | False | False | False | False |
Let’s suppose we’re dealing with binary data. We convert all value to binary, in order to be able to compare them correctly.
The cell below needs refactoring for different type of data. In case of nominal data inside strings it might be enough to skip the cell below. A custom solution is needed for more complex use cases. Additionally, make sure to adopt the majority decision cells to any changes down here and vice versa.
# Function to convert a column to boolean if it's not already
def convert_to_bool(column):
if contingency_table[column].dtype != 'bool':
bool_map = {'True': True, 'False': False}
return contingency_table[column].map(bool_map)
return contingency_table[column]
# Convert columns to boolean if they are not already
contingency_table['Majority Decision']= convert_to_bool('Majority Decision')
contingency_table['Model'] = convert_to_bool('Model')Let’s quickly check pairwise agreement between the Model and Majority Decision using Cohen’s Kappa:
kappa_scores = calculate_kappa(contingency_table[['Majority Decision', 'Model']])
pd.DataFrame(kappa_scores).head()| Coder 1 | Coder 2 | Kappa | Overlap | Coding | |
|---|---|---|---|---|---|
| 0 | Majority Decision | Model | 0.831461 | 50 | Positioning |
Machine Learning Metrics
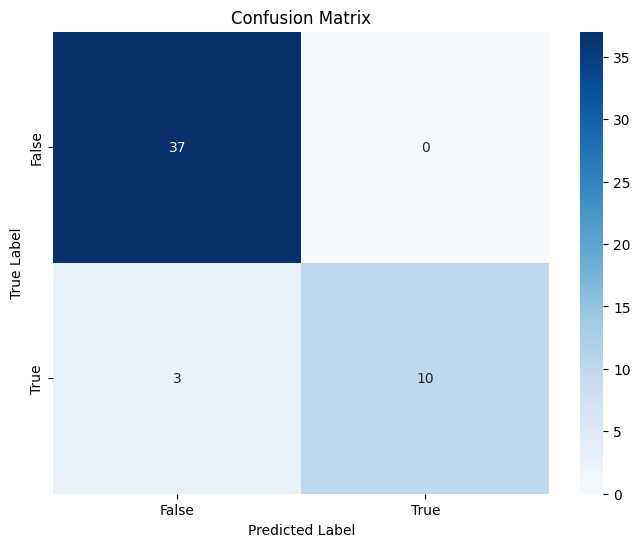
Finally, let’s calculate Accuracy, Precision, and F1 Score and plot a confusion matrix.
import pandas as pd
from sklearn.metrics import accuracy_score, precision_score, recall_score, f1_score
from IPython.display import display, Markdown
# Calculating metrics
accuracy = accuracy_score(contingency_table['Majority Decision'], contingency_table['Model'])
precision = precision_score(contingency_table['Majority Decision'], contingency_table['Model'], average='binary')
recall = recall_score(contingency_table['Majority Decision'], contingency_table['Model'], average='binary')
f1 = f1_score(contingency_table['Majority Decision'], contingency_table['Model'], average='binary')
# Creating a DataFrame for the metrics
metrics_df = pd.DataFrame({
'Metric': ['Accuracy', 'Precision', 'Recall', 'F1 Score'],
'Value': [accuracy, precision, recall, f1]
})
# Displaying the DataFrame as a table
display(metrics_df)| Metric | Value | |
|---|---|---|
| 0 | Accuracy | 0.940000 |
| 1 | Precision | 1.000000 |
| 2 | Recall | 0.769231 |
| 3 | F1 Score | 0.869565 |
import seaborn as sns
from sklearn.metrics import confusion_matrix
import matplotlib.pyplot as plt
# Generate the confusion matrix
cm = confusion_matrix(contingency_table['Majority Decision'], contingency_table['Model'])
# Plotting the confusion matrix
plt.figure(figsize=(8, 6))
sns.heatmap(cm, annot=True, fmt='d', cmap='Blues', xticklabels=['False', 'True'], yticklabels=['False', 'True'])
plt.title('Confusion Matrix')
plt.xlabel('Predicted Label')
plt.ylabel('True Label')
plt.show()